Understanding the structure, function, and three-dimensional (3D) architecture of the neuronal tissue relies heavily on imaging technologies. During the last decades, confocal (CF) microscopy constituted the standard fluorescence imaging technology for neuronal tissue analysis. Although this approach can be sufficient to study cellular and tissue morphology and dynamics, sometimes it may not be enough to accurately answer all biological questions.
In this context, Super-Resolution (SR) microscopy is becoming an increasingly popular and robust approach across neurobiological research, and life science in general, to study fine cellular structures and subcellular components. This further level of information could allow new discoveries to be made and reveal details otherwise hidden by the diffraction limit.
In this Application Note, we demonstrate the great flexibility of the CrestOptics X-Light V3 CF system, coupled with DeepSIM X-Light SR system, for combining different imaging modalities and covering a wide range of neurobiological applications.
A comprehensive overview of complex brain structures can be obtained by combining large field-of-view CF acquisitions and SR imaging
The ongoing quest to map the intricate brain’s cytoarchitecture with increasing accuracy and resolution is expanding in new directions of imaging solutions. Here we propose three examples of CF and SR imaging combinations to resolve intricate networks of cellular circuits in the mouse brain.
For this purpose, we used a set-up consisting of X-Light V3 CF spinning disk system coupled with DeepSIM X-Light SR system; two different technologies sharing the same light source, camera, emission and excitation optics and, the sample itself.
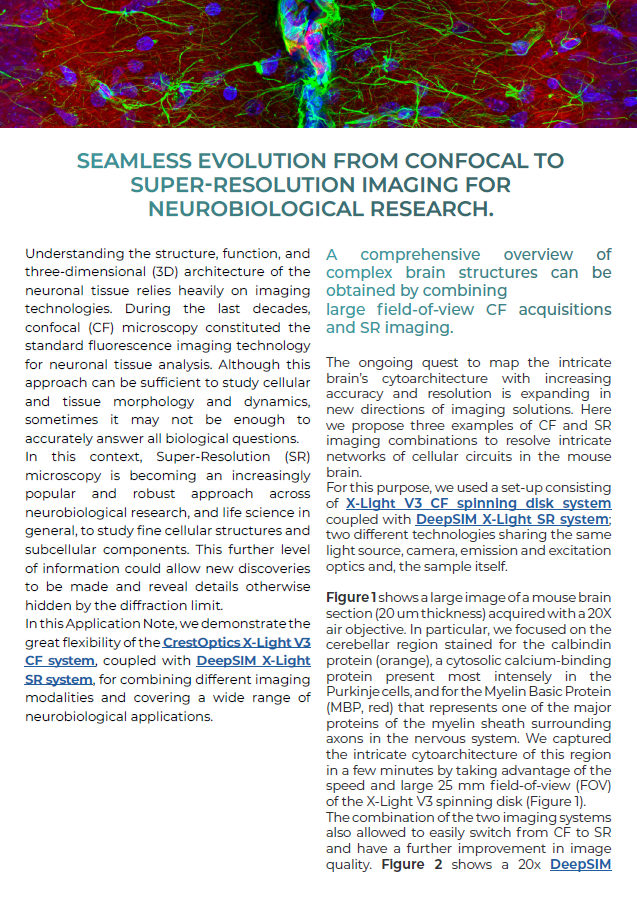
Figure 1 shows a large image of a mouse brain section (20 um thickness) acquired with a 20X air objective. In particular, we focused on the cerebellar region stained for the calbindin protein (orange), a cytosolic calcium-binding protein present most intensely in the Purkinje cells, and for the Myelin Basic Protein (MBP, red) that represents one of the major proteins of the myelin sheath surrounding axons in the nervous system. We captured the intricate cytoarchitecture of this region in a few minutes by taking advantage of the speed and large 25 mm field-of-view (FOV) of the X-Light V3 spinning disk (Figure 1).
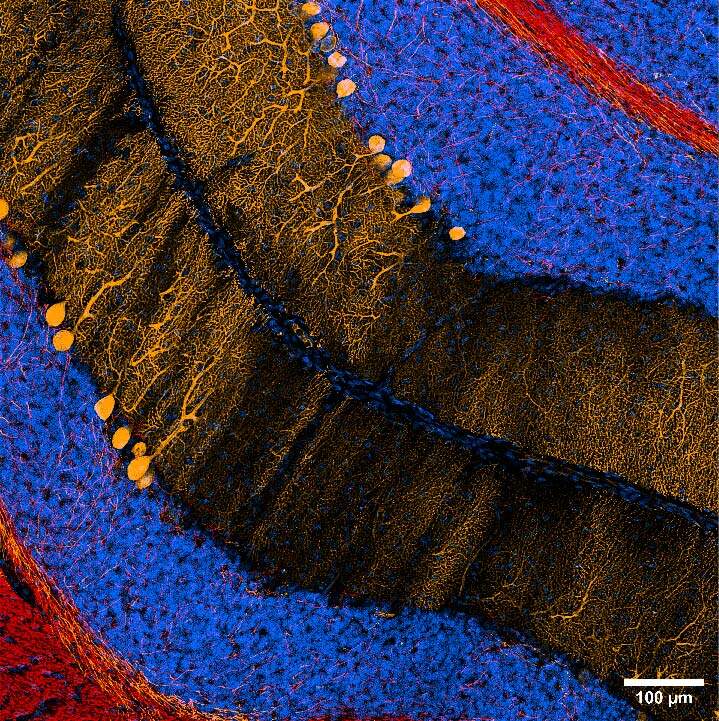
The combination of the two imaging systems also allowed to easily switch from CF to SR and have a further improvement in image quality. Figure 2 shows a 20x DeepSIM stitching of a large portion of the cerebellum and a 60X Z stack (20um maximum intensity projection), revealing the intricate network of neuronal connections covered by myelin.
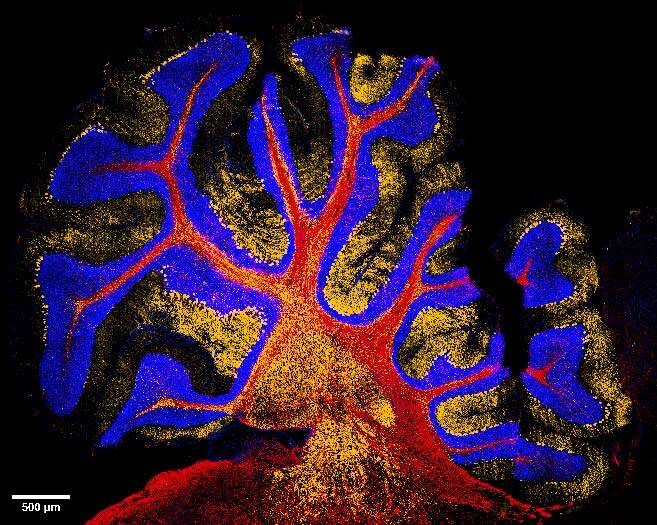
Figure 1: Large image of mouse cerebellum stained for MBP (red) and calbindin (orange); nuclei are stained in blue. Scale bar: 500 um. This image was acquired with a CrestOptics X-Light V3 spinning disk equipped with a Nikon CFI Plan Apochromat Lambda D 20X air objective.
A
B
Figure 2: (A) Stitching of mouse cerebellum stained for MBP (red) and calbindin (orange); nuclei are stained in blue. Scale bar: 100 um. This image was acquired with a CrestOptics DeepSIM SR system equipped with a Nikon CFI Plan Apochromat Lambda D 20X air objective. (B) Maximum intensity projection from 20 um Z stack of neuronal network displaying MBP positive myelin sheath (red) and calbindin positive neurons (orange). Scale bar: 20 um. This image was acquired with a CrestOptics DeepSIM X-Light SR system equipped with a Nikon CFI Plan Apochromat Lambda D 60X oil objective.
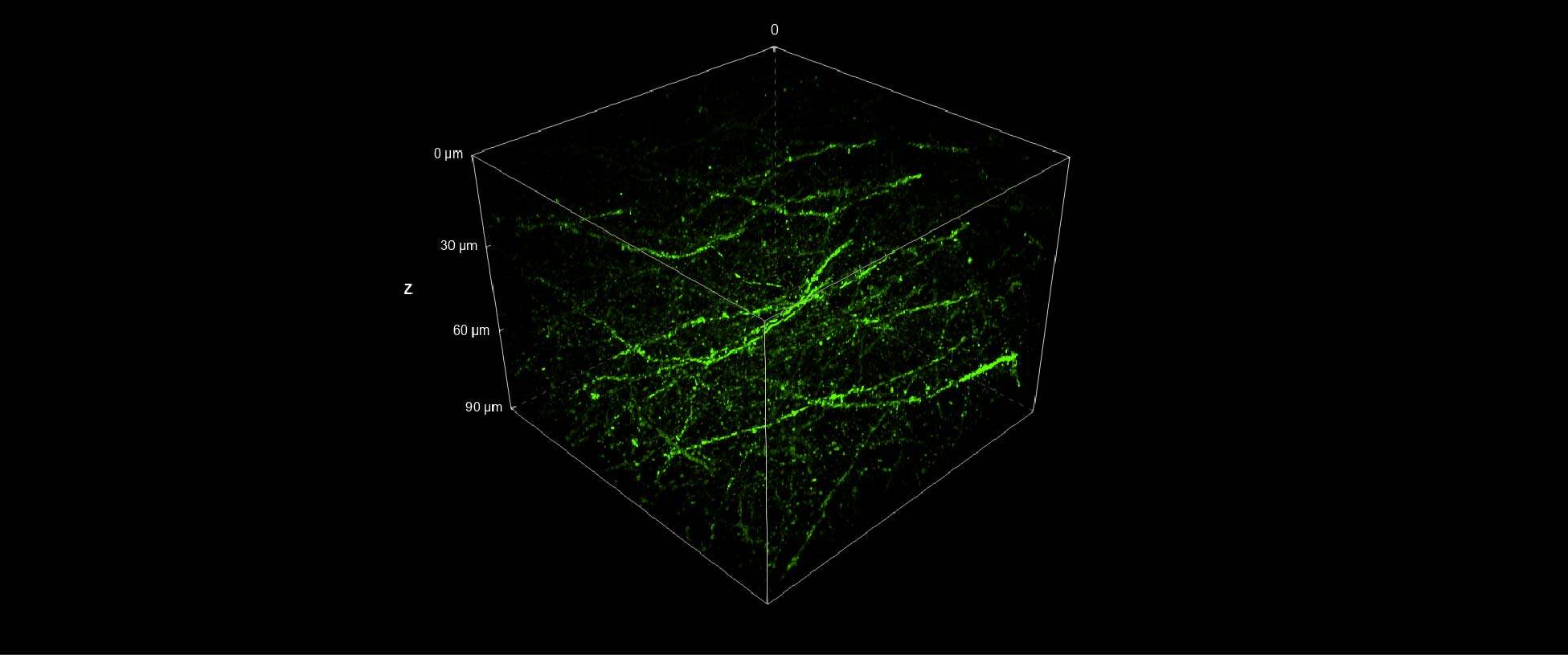
Furthermore, we applied the same strategy of combining the stitching capacity on large FOV of the V3 with the resolution potential and penetration depth of the DeepSIM to acquire a whole cleared mouse brain section (250 um thickness) with GFP-expressing neurons. The entire mouse brain coronal section (43 mm2 area) was acquired using the CrestOptics X-Light V3 CF spinning disk system (Figure 3) in a few minutes. By having such a wider FOV, more information can be collected, and fewer tiles are needed to cover a large sample, enabling the imaging process to be significantly accelerated. Next, we switched to the DeepSIM X-Light SR system and we focused on dendritic spines, nanometer-sized protrusions along the dendritic shaft, in the cortical region of the mouse brain. As demonstrated in Figure 4, the SR system was crucial to enhance the details of such fine biological structures even within a very thick sample; both 90 um 3D view (Figure 4A), and maximum intensity projection (Figure 4B), clearly showed dendritic spines.
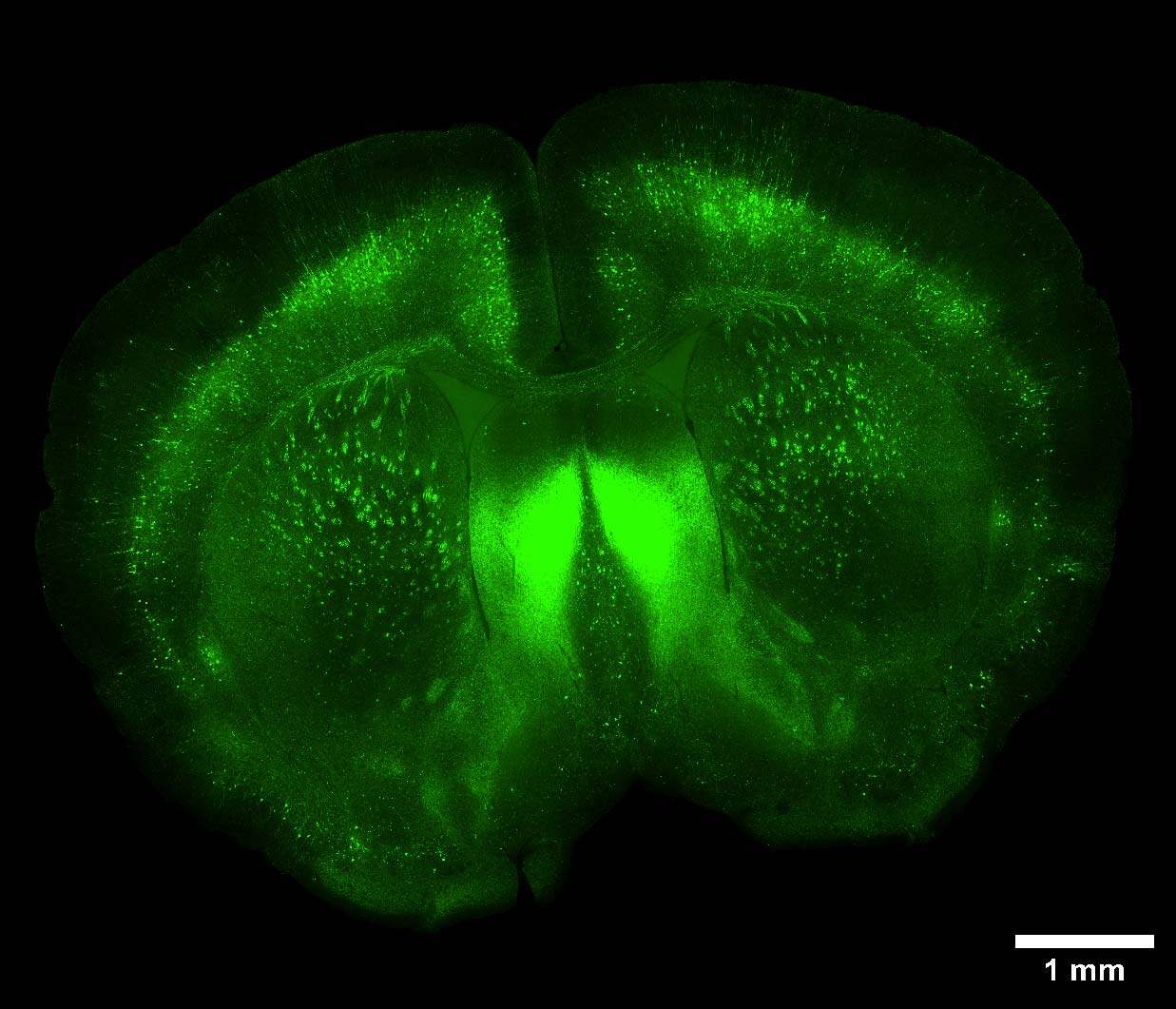
Figure 3: Large image of a whole cleared mouse brain section with GFP-expressing neurons. Scale bar: 1 mm. This image was acquired with a CrestOptics X-Light V3 spinning disk equipped with a Nikon CFI Plan Apochromat Lambda D 20X air objective.
A
B
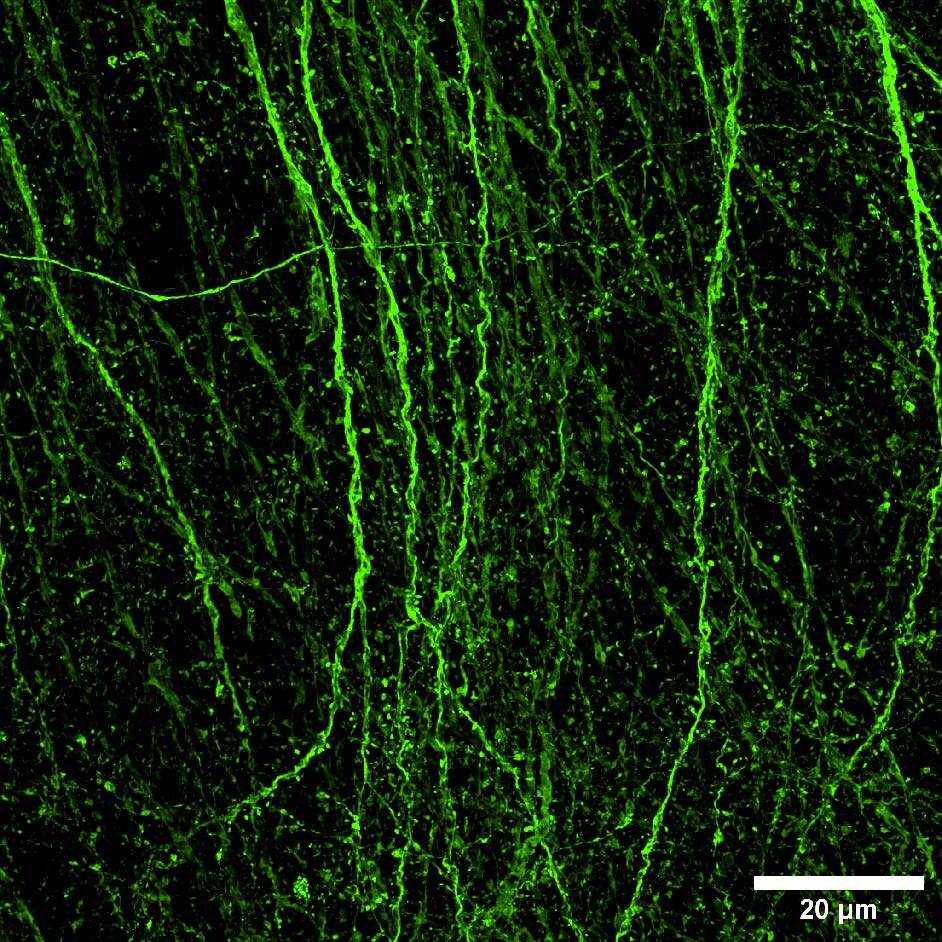
Figure 4: (A) DeepSIM SR 3D volume view of 90 um Z stack. (B) Maximum intensity projection from 90 um Z stack. Scale bar: 20um. This image was acquired with a CrestOptics DeepSIM X-Light SR system equipped with a Nikon CFI Plan Apochromat Lambda D 60X oil objective.
The latest example of combining large FOV CF acquisitions and SR imaging is shown here for the visualization of reactive astrogliosis in a mouse brain section. Reactive astrogliosis is characterized by a profound change in astrocyte phenotype in response to all CNS injuries. Glial fibrillary acidic protein (GFAP) is an intermediate filament protein found in the astroglial cytoskeleton and, for its remarkable specificity, it is recognized as a classical biomarker for astrocytes and traumatic brain injury (TBI).
In Figure 5 a perfect stitching of a mouse brain slice (21 mm2 area) with a GFAP staining is shown. This acquisition provides an excellent starting point for gaining a general overview of the sample and assessing the reactivity of the astrocytes on the whole section.
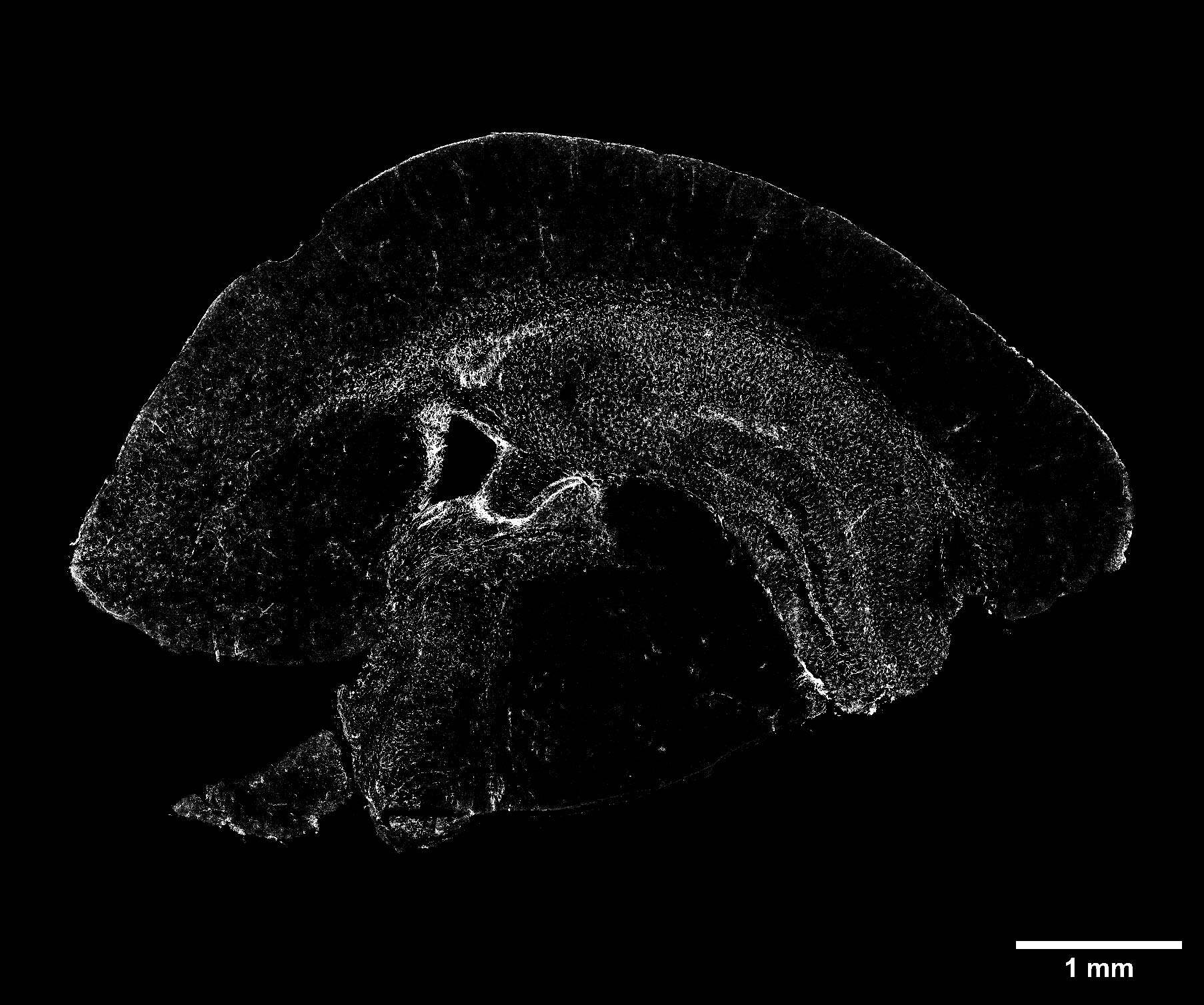
Figure 5: Large image of a whole mouse brain section stained for GFAP-positive astrocytes (white). Scale bar: 1 mm. This image was acquired with a CrestOptics X-Light V3 spinning disk equipped with a Nikon CFI Plan Apochromat Lambda D 20X air objective.
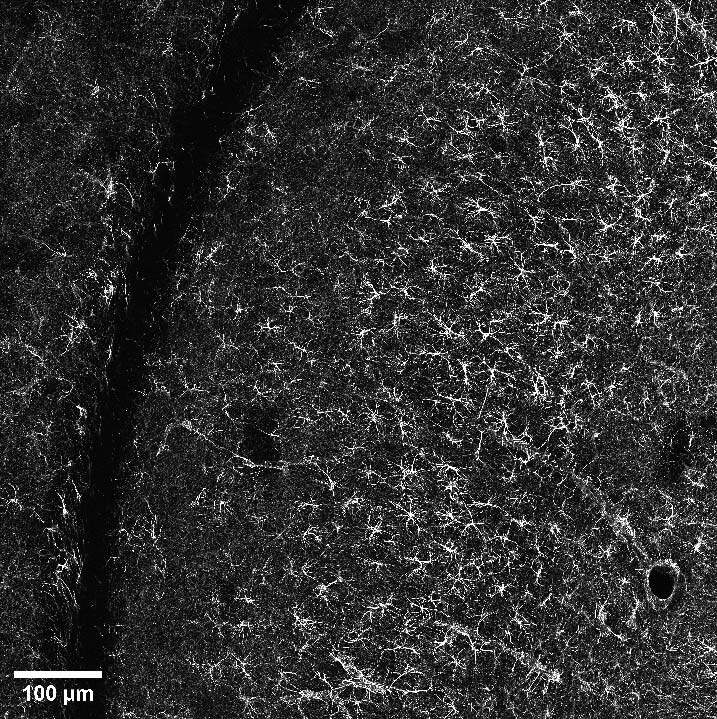
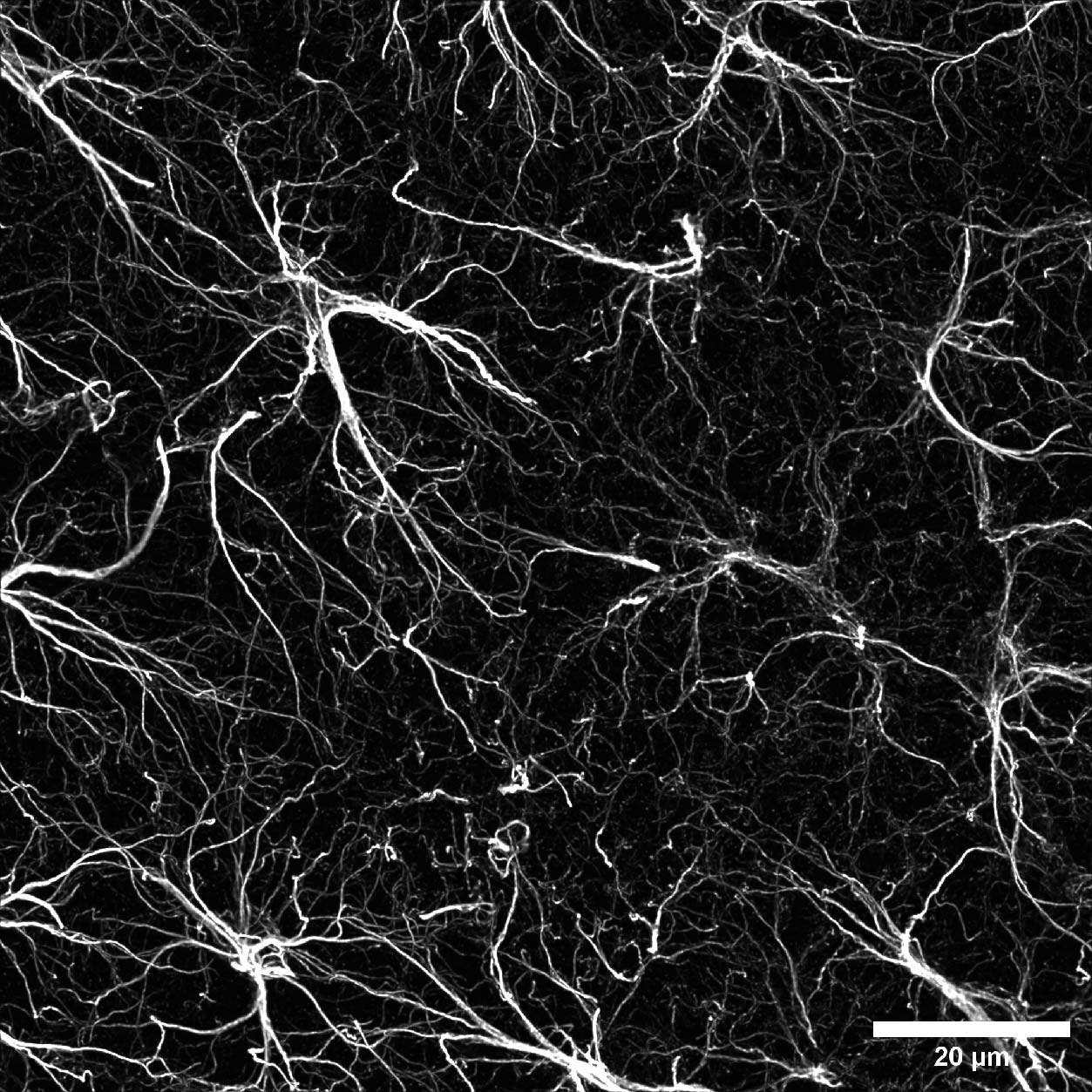
Once the area of interest was found, it was possible to gradually increase the resolution by selecting the SR mode. In particular, Figure 6A represents a DeepSIM, stitching acquired with a 40X water objective of a mouse brain large area; on the other hand, a maximum intensity projection of 40 um Z stack acquired with a 60X oil objective is shown in Figure 6B. Finally, we isolated a single astrocyte to study its morphology in 3D. The 3D video shown in Figure 6C demonstrates the ability of the DeepSIM to work even with thick samples, enabling single-cell analyses and studying astrocyte morphology under physiological and pathological conditions.
A
B
C
Figure 6: (A) Stitching of mouse brain stained for GFAP astroglial marker. Scale bar: 100 um. This image was acquired with a Nikon CFI Apo Lambda S 40XC WI objective. (B) Maximum intensity projection from 40 um Z stack of mouse brain section displaying GFAP-positive astrocytes. Scale bar: 20 um. This image was acquired with a Nikon CFI Plan Apochromat Lambda D 60X oil objective. (C) 3D movie showing a single astrocyte shape. All three images were acquired with a CrestOptics DeepSIM X-Light SR system.
SR imaging reveals membrane-associated periodic cytoskeleton structures in neuronal axons
Cytoskeleton organization and dynamics play an essential role in polarization, synaptogenesis, axon growth, and degeneration of neurons. The actin protein is responsible for the formation of microfilaments in the cytoskeleton of cells; in neurons, actin was shown to form ring-like structures around the axonal circumference with a periodic spacing of 180-190 nm. These neuronal actin rings contain adducin, an actin-capping protein, and are crosslinked by spectrin tetramers that contribute to the spacing between individual rings (Figure 7). A periodic lattice of spectrin tetramers forms a highly stable structure that provides mechanical support for the thin structure of axons.
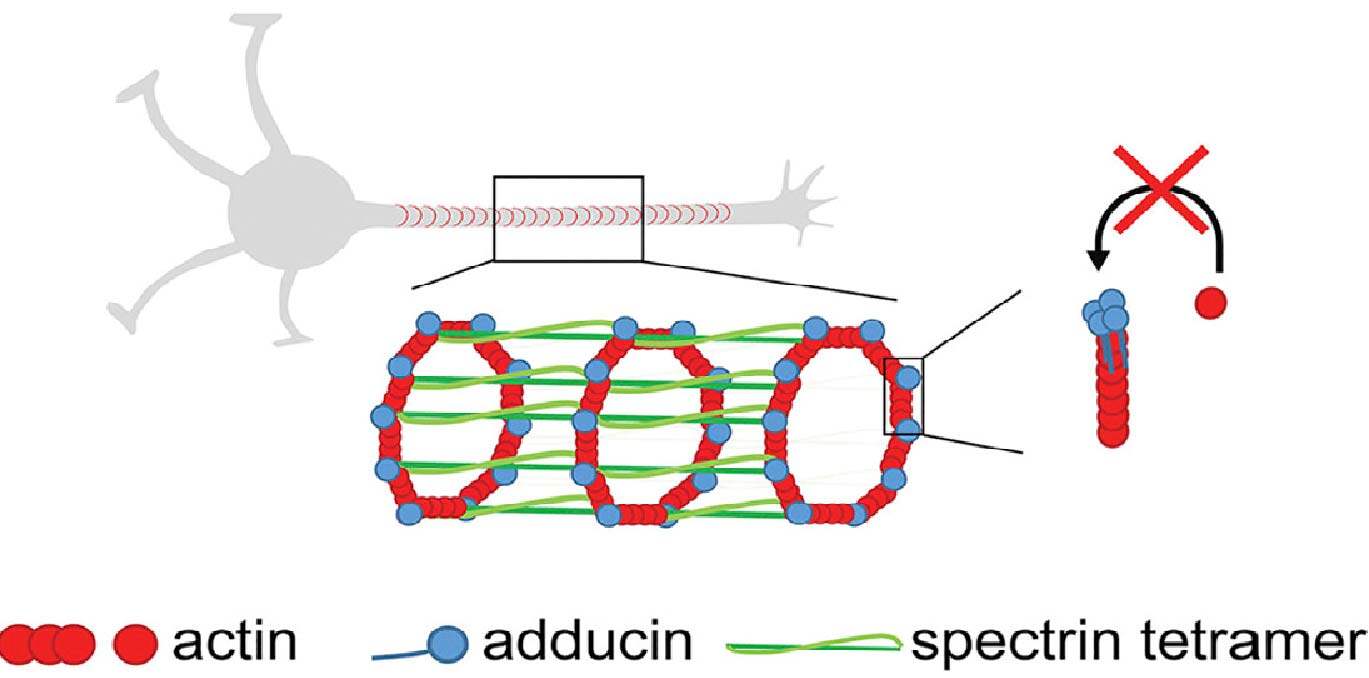
Figure 7: Ring-like structure around axonal circumference composed of actin (red), adducin (blue) and spectrin tetramer (green). This image was adapted by Leite et al., 2016 under the terms and conditions of the Creative Commons Attribution license (CC BY 4.0).
Since membrane-associated proteins have a periodic distribution around 190 nm, it is clear that CF imaging cannot visualize these structures below the diffraction limit. Here we demonstrate that with the use of the DeepSIM, coupled with a high NA objective (100X, 1.45 NA), it is possible to reveal these membrane-associated 190 nm periodic cytoskeleton organization.
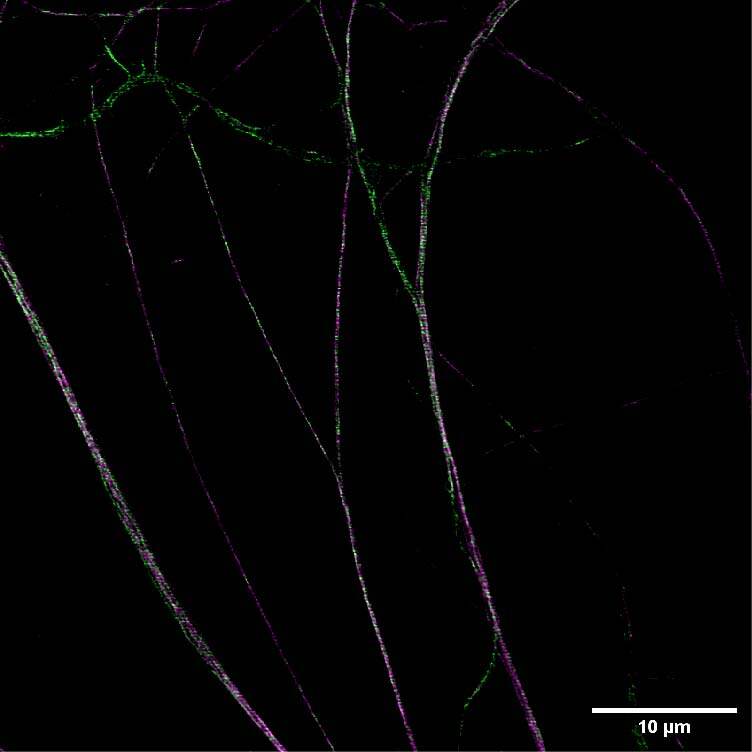
We acquired some axonal regions of a culture of primary mouse neurons (Figure 8A), stained for adducin (green) and β2 spectrin (magenta), and then we focused on the middle regions of the axons, where the membrane-associated periodic structures are most periodic. Figure 8B demonstrates that the impressive increase in resolution allowed by DeepSIM allowed us to visualize the structural alternation of adducin and spectrin; these structures can only be glimpsed in a CF image (Figure 8B, left).
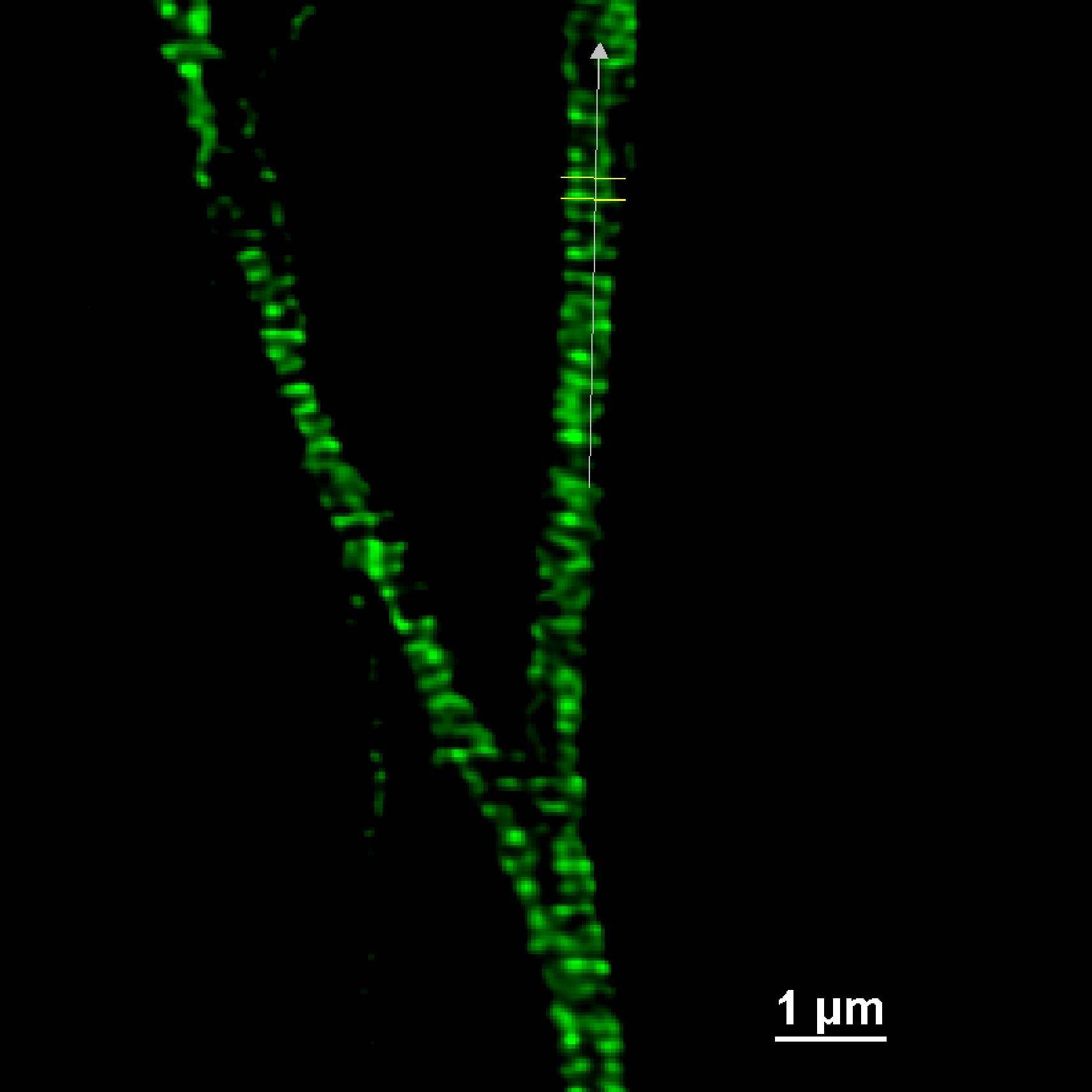
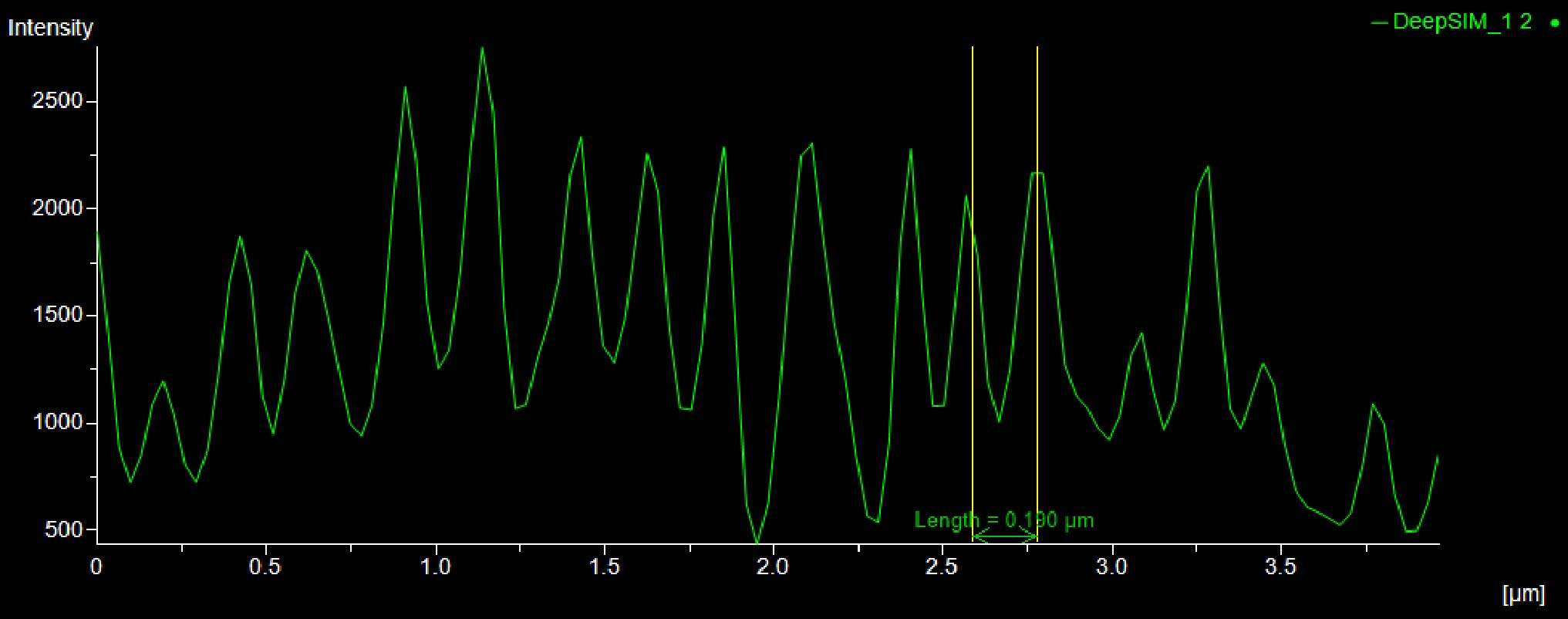
Moreover, as shown in Figure 9, the distance (um) between the peaks of the intensity profiles of both the adducin (Figure 9A) and spectrin (Figure 9B) structures demonstrates the 190 nm periodicity of this architecture.
According to these results, the DeepSIM X-Light SR system revealed periodic cytoskeleton structures associated with membranes in neuronal axons, thus setting up the possibility of observing and analyzing biological structures at the subcellular level.
A
B
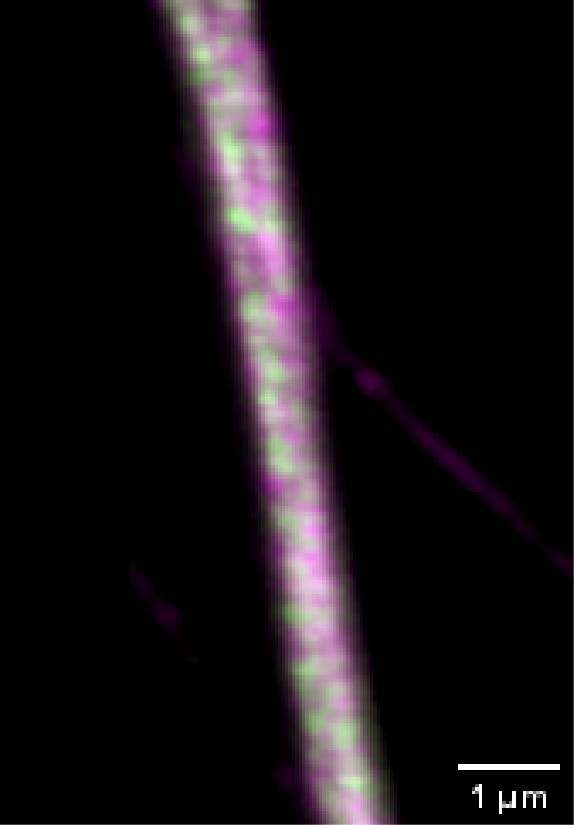
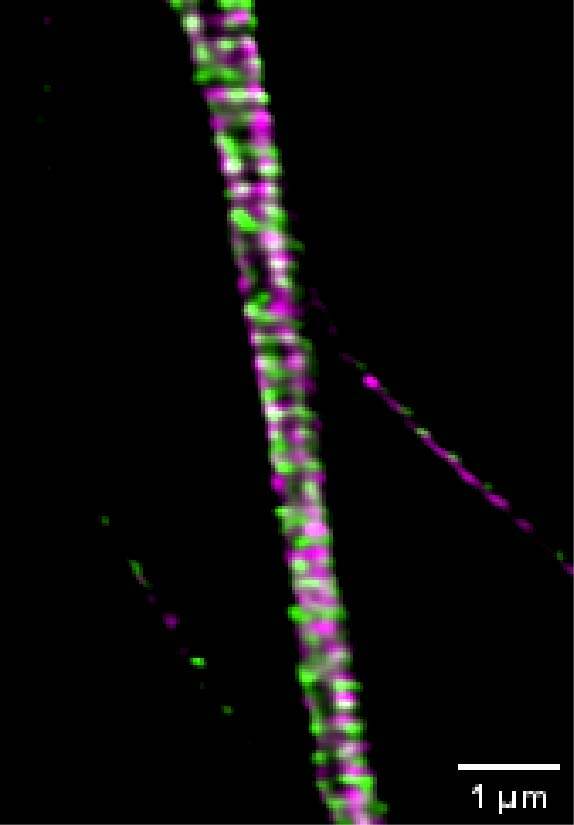
Figure 8: (A) SR acquisition of axonal regions of a culture of primary mouse neurons stained for adducin (green) and β2 spectrin (magenta). Scale bar: 10 um. (B) Comparison of CF (left) and SR (right) of a middle region of a neuronal axon. Scale bar: 1 um. These images were acquired with a CrestOptics DeepSIM X-Light SR system equipped with a Nikon CFI Plan Apochromat Lambda 100X oil objective.
A
B
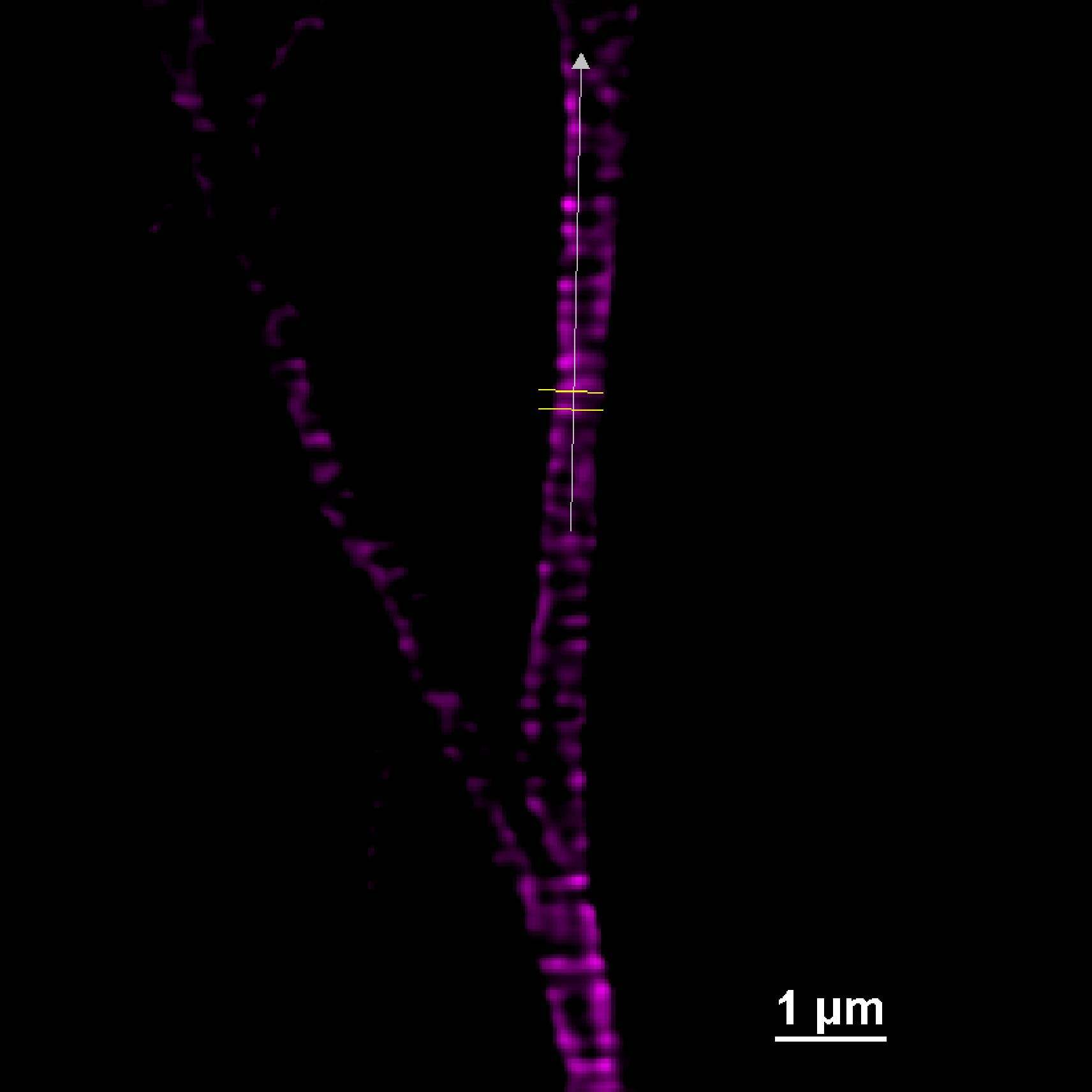
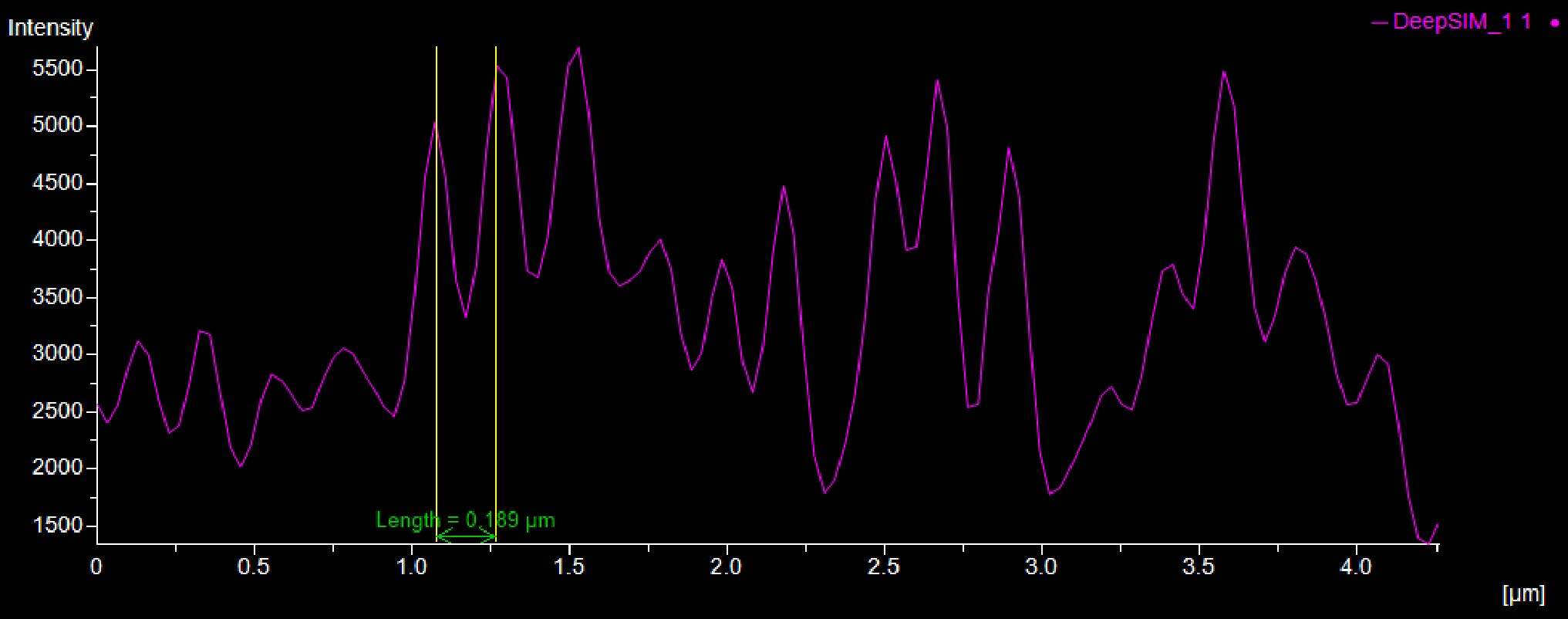
Figure 9: SR acquisition, and intensity profiles, of axonal regions of a culture of primary mouse neurons stained for adducin (A, green) and β2 spectrin (B, magenta) and . Scale bar: 1 um. These images were acquired with a CrestOptics DeepSIM X-Light SR system equipped with a Nikon CFI Plan Apochromat Lambda 100X oil objective.
Conclusion
Altogether these data demonstrate the great flexibility of the CrestOptics X-Light V3 CF system, coupled with the DeepSIM X-Light SR system, for combining different imaging modalities in neurobiological studies.
In particular, the capacity to image a FOV up to 25 mm, combined with the homogeneous illumination, makes the X-Light V3 the perfect solution for the seamless stitching of large samples. Customers can benefit from the extreme speed of the system and obtain a complete overview of the sample in a short time.
Furthermore, the system is compatible with the DeepSIM X-Light SR system, for a seamless evolution from CF to SR. Where an increase in resolution is needed, our customers can easily switch to SR mode and focus on a specific region of the sample to reveal biological structures below the diffraction limit. A two-fold increase in spatial resolution can be obtained using both high magnification (60X, 100X) and low magnification (20X, 40X) objectives, thereby enabling the study of complex 3D neurobiological tissues.
In conclusion, here we proposed the CrestOptics X-Light V3 CF system, coupled with the DeepSIM X-Light SR system, as a modular, expandable, extremely flexible, and highly performant system to cover a wide range of neurobiological applications and beyond.
Acknowledgments
We thank Dr. Antonio Virgilio Failla and Dr. Bianka Brunne, Universitätsklinikum Hamburg-Eppendorf UKE Microscopy Imaging Facility (UMIF), and Dr. Ruobo Zhou, professor at The Pennsylvania State University, for kindly providing some of the samples shown in this application note.
The image in Figure 7 was adapted by Leite et al., 2016 under the terms and conditions of the Creative Commons Attribution license (CC BY 4.0).